OverExpressed
OverExpressedEagerly awaiting my genetic destiny
I’m about to see some things that can’t be unseen, to learn some things that can’t be unlearned, and to think some things that can’t be unthought. Within less than a fortnight, a significant amount of my genetic destiny will be revealed to me by the magic of consumer genetics. And the suspense is killing me.
As a metrics and diy bio junky, I’ve been very eager to explore the essential blueprints to my being. It’s incredible to even imagine that most everything I am can be reduced to a fundamental set of instructions based on patterns of just four letters (the rest can be accounted for by ‘nurture’, which for me is mainly an amalgam of adventure novels and Saved by the Bell episodes, as best I can tell).
When I first heard about 23andme several years ago, I was pretty sure the future was coming fast. However, the price tag was still a bit steep for me. Considering the Moore’s Law depreciation of sequencing costs, I just couldn’t rationalize the expense for a report that didn’t even cover my entire genome. I even signed up for the Personal Genome Project in the interim. Unfortunately, they still haven’t taken me in, and my enrollment seems unlikely given their preference for people with known rare genetic conditions. So I’ve waited for the price to go down.
Happy DNA Day!
And finally an opportunity for low-cost genotyping! April 23rd was National DNA Day, during which 23andme (named after your 23 chromosome pairs), offered their full package (including ancestry, health, and extended sequence access) for just $99. This was a $400 discount from the normal $499 price tag. Obviously, I jumped on the deal.
Of course, 23andme is just one of many consumer sequencing companies (including Navegenics, deCODEme, and Knome, among others). However, 23andme offers one of the most complete offerings I’ve found. They give you ~600,000 known “single nucleotide polymorphisms” (or SNP’s, basically just single letters in your sequence where you’re likely to vary from others in a meaningful way), including mitochondrial DNA. Granted, this is just a small portion of my entire genome (only ~0.02% of my 3 billion bases, to be ~exact). However, it represents many of the significant places (or loci) where I differ from you or anyone else. It also includes many compelling factors involved with a range of heritable conditions. And I’ll be particularly interested to learn things like my eye and hair color.
Some Reasonable Caution
 Now, it’s not a terrible idea to take a step back and consider the consequences of such deep self-knowledge. First of all, I have to ponder the psychological impacts this information could have on me. What if I find out I have some rare genetic disorder that is reliably linked to a terminal illness? What if I find some factors that would indicate the need for a drastic change in lifestyle? What if my dad isn’t my dad? What if I find that I’m completely boring, genetically? These are all possibilities. But I’m prepared (or at least momentarily indifferent) to their consequences. I believe that understanding the root of your medical condition can help you make educated choices moving forward, and I intend to leverage any information I gain.
Now, it’s not a terrible idea to take a step back and consider the consequences of such deep self-knowledge. First of all, I have to ponder the psychological impacts this information could have on me. What if I find out I have some rare genetic disorder that is reliably linked to a terminal illness? What if I find some factors that would indicate the need for a drastic change in lifestyle? What if my dad isn’t my dad? What if I find that I’m completely boring, genetically? These are all possibilities. But I’m prepared (or at least momentarily indifferent) to their consequences. I believe that understanding the root of your medical condition can help you make educated choices moving forward, and I intend to leverage any information I gain.
However, I am slightly more suspicious of the potential legal implications. Just imagine a Gattaca-esque world (by the way – notice the title is made of A, T, G, C) where your job, insurance, and even mate, are essentially determined by the strength of your genetic code. That’s some pretty scary stuff. And most people don’t seem to realize how close we are to this (at least technologically). There are already a number of opportunities for genetic theft. It will be very important for the government to enact some tough regulations that can withstand any assault on personal genomic privacy. Fortunately, we currently have a law protecting us from genetic discrimination with respect to insurance and employment.
What I Hope to Get Out of This
It’s no secret that I’m a fan of data. I’d like to be as quantitative as possible in my life choices. Within the realm of health, this has primarily manifested itself in the use of activity-tracking applicaitons, such as CardioTrainer for running and Daily Burn (formerly Gyminee) for weight training. However, my genome represents a vast bank of data that I could never empirically derive by phenotypic analysis alone. I’m super excited to finally get access to even a small portion of this raw data.
Although obviously I could have accessed a lot of this information by myself using several methods (including outsourcing directly to Illumina, as 23andme has done), I wanted to go through one of these companies (and 23andme in particular). This is mainly because I appreciate all of the additional analysis and formatting they provide in presenting the absolutely daunting amount of information contained in my raw genetic sequences. They’ve developed some simple tools to show my ancestry, as well as my health risks (weighted by the reliability of the associated studies in a 5-star format) in a secure, web-based format. Though I could collect and analyze this information myself, it would certainly take a significant amount of effort, and thus I’m willing to pay a service to provide this convenient user experience.
And speaking of user experience, it’s interesting to note the relationship between 23andme and Google. Specifically, 23andme received a significant amount of funding from Google (mildly controversial since 23andme cofounder Anne Wojcicki is married to Google cofounder Sergey Brin). But this all makes sense to me. It is Google’s stated goal to “organize the world’s information and make it universally accessible and useful.” Personal genomic data is perhaps some of the most useful information out there, and Google clearly has an interest in organizing it. Considering what they’ve done for the web, I’m excited to see what they can do to simplify genetic relationships. And I suppose I’m about to find out. All my base are belong to Google.
Continue Reading | No Comments
Tags: 23andMe, biology, Consumer Genetics, DNA, Genetics, Knome, Personal Health, Science, Single-nucleotide polymorphism
The Scientific Journal of Failure
Science is riddled with failure
Really, it’s all over the place. It’s built right into the scientific method. You make a hypothesis, with a firm understanding that anything could happen to disprove your faulty notions. Sometimes it works and you see what you expected, and sometimes it doesn’t. And some of the most interesting discoveries of all time have come from these “accidents” where researchers stumbled on something that didn’t work how they expected it to.
However. The majority of these scientific snafus result in absolutely nothing. You just blew 16 hours and $3,000. Gone. But at least now you know that mixing A & B only produces C when you have the conditions very precisely set at D. But what happens to all of your failed attempts? Your (very expensive) failed attempts?
They disappear. There is neither forum nor incentive for researchers to publish their failures. This leads to an enormous amount of redundant effort, burning through millions of dollars every year (many of which are funded by the tax-payer). The system sounds really inefficient, right? So why do we do it?
People tend to focus on successes. Sure, things didn’t work out 6 or 7 or 17 times…but then one time it all came together. And then you go back and repeat every single obscure condition that led to your ephemeral success (even wearing the same underwear and eating the same breakfast – but maybe I’m just more scientifically rigorous than most). And what do you do? You manage to repeat the success and you publish it. Maybe you publish a parametric analysis showing how you optimized your result. But you have left a multitude of errors and false starts in your wake that will never see the light of day, leaving countless other researchers to stumble into the very same unfortunate pitfalls. It’s a real waste.
But this protects you. In the business world they call it “barriers to entry“. You’ve managed to find yourself racing ahead in first place on something and the only thing you can do to protect yourself is drop banana peals to trip up your competitors (fingers crossed nobody gets a blue turtle shell). On top of that, every time you publish something, you’re putting your reputation on the line. A retraction can be devastating, particularly for an early career. Why take such a risk just to publish something that doesn’t even seem very significant? It doesn’t help you any. But is this good for science? Of course not. You’re delaying progress. You’re wasting money. And, particularly in the medical sciences, you’re probably actually killing people.
So Let’s Document The Traps

I propose a journal devoted entirely to failures. Scientists can publish any negative results that are deemed “unworthy” of standard publication. Heck, we can even delay publication by 6-12 months, to give the authors a little head-start on their competitors. And there would have to be some sort of attribution. But nobody wants their name attached to The Journal of FAIL. So let’s call it The Scientific Journal of “Progress”. Maybe without the quotes.
I wasn’t sure if such a journal existed already (though if it does, I certainly haven’t heard of it – and I’d probably be one of the biggest contributors, right behind this guy). The only close option that came up was the Journal of Failure Analysis and Prevention, but they seem to be focused on mechanical/chemical failures in industrial settings. So the playing field is still wide open.
Imagine if every experimental loose end was captured, categorized, and tagged in an efficient manner so that any person following a similar path in the future could actually have a legitimate shot at learning something from the mistakes of others. It would be kind of like a Nature Methods protocol, but actually including all of the ways things can go wrong. Authors could even establish credibility by describing why certain conditions didn’t work (a very uncommon practice among scientists). We might have to develop some technological tools that automatically pull and classify data as researchers collect it, making it a simple “tag and publish” task for the researcher. And people could reference your findings in the future, providing you with even more valuable street cred for the entire body of work you have developed (not just the sparse successes).
Of course none of this will happen any time soon. There simply isn’t enough of a motivating force to drive this kind of effort (besides good will). Maybe one day science will get its collective head out of its collective ass and things will change. And this will be one of those things. In the meantime, I will do my best to record and post tips on avoiding my own scientific missteps.
BONUS WEB2.0 TOOL: TinEye
[you’ve read this far, you deserve a reward]
I originally did a google search for “Science Fail” and found the first image of this post on another blog. I was curious about the actual origins of the image, so I used the TinEye image search engine to find the real source. Tineye is just like any other search engine, except your query is an actual image. You just upload (or link to) an image of interest, and TinEye scours the web for any similar images (actual pixels, not metadata). I quickly found that the picture originally camed from a “Science as Art” competition sponsored by the Materials Research Society (MRS). TinEye searched over 1.1011 billion images in 0.859 seconds to find that result. Pretty nifty, right?
The obvious applications are for controlling the distribution of copyrighted images, and there are some potentially frightening applications of such technology for facial recognition in the not-so-distant future (imagine someone sees you on the street, snaps a picture surreptitiously, and does a quick image search to find out who you are – and they’re likely to find out everything). Scary. But still kind of fun.
Tags: Image Search, Medicine, Publications, Scientific Journal, Scientific method
Become a Research Subject
Rick's Brain on Science
Admit it. You like science. You thrive on the unknown. You seek adventure and mystery. You enjoy being enclosed in absurd magnetic fields while grad students sit safely in the room next door. Or maybe I’m alone there. But at least you like money.
Most universities with research programs have science. And that science sometimes needs human subjects. And usually those human subjects aren’t really subjected to anything that would be considered inhumane (as opposed to the kinds of things that inhuman animals are subjected to). And sometimes, if you’re lucky, those human subjects can be you.
There are many human experiments out there that are really quite mild (ask a few questions, fill in a few surveys, watch a few videos, spit into a cup, etc.), but they’re pretty interesting and can pay up to $15-$50 per hour for your trouble. Sure, you’re not going to make a living off of it (though some have, and there is a legitimate debate over a minimum wage for guinea pigs), but it’s a nice diversion that can help you rationalize buying a new wetsuit (true story).
So how do you get involved? If you live in Berkeley, just sign up for the Research Subject Volunteer Program (RSVP) and you can participate in all kinds of cool studies (mostly involving psychology – so it’s just fun games/questions). On top of that, if you do any kind of MRI experiment, you’re very likely to get a killer picture of your brain (as shown above). If you’re not fortunate enough to find yourself in the bay area, I would recommend doing a quick google search for “Research Subject Volunteer” + “Your Local Big University” and you’re likely to find some results.
So far I’ve participated in a few experiments between Berkeley and UCSF. I played some memory games in an MRI machine and got a picture of my brain (I thought the machine was relaxing and even started to fall asleep – my roommate, on the other hand, was intensely uncomfortable and vowed to never again volunteer for unnecessary cranial imaging). I’ve also watched some violent videos, to which my responses are presumably being correlated with genetic information found in my spit. So it’s all cool stuff.
They even have an experiment that will give you $210 to play video games for a month:
Relationships and Social Cognition Lab emotion and video games Three part study, involves two laboratory visits during which physiological responses will be monitored. In between lab visits participants will be asked to play computer games daily for half an hour for 30 days. Compensation: up to $210 Special Requirements:completed high school in the US, fill out screener: http://www.surveymonkey.com/s.aspx?sm=GWjkpIDz5amM1XaOd_2fZZ8A_3d_3d Location: UC Berkeley Campus
An interesting aside: in researching this article (yes, there was some “research” involved), I found that the Tufts School of Dental Medicine is the first result in a google search for “become a research subject”. I just particularly enjoyed how they “broke it down” for the laypeople:
What Is Research?
- Research is a study that is done to answer a question.
- Scientists do research because they don’t know for sure what works best to help you.
- Some other words that describe research are clinical trial, protocol, survey, or experiment.
- Research is not the same as treatment.
Maybe, just maybe, your diggs and reddits and tweetiemabobs will push the google-rated significance of this article above those tooth jockies at Tufts. Yes We Can!
Little-known fact: I do it for the adventure and mystery.
Continue Reading | No Comments
Tags: Psychology, Research, University of California Berkeley, University of California San Francisco
Google has a problem with DNA analysis
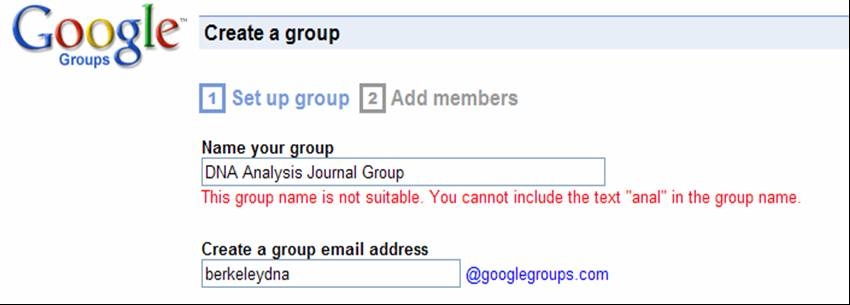
About a year ago, my roommate started a little DNA analysis journal club here at Berkeley. It was just meant to be a group of like-minded students discussing recent advances in analytical DNA technologies. He tried creating a Google group for that club, not expecting the fairly judgmental response he received…
Note that Google’s skilled text parser caught our hidden innuendo:
This group name is not suitable. You cannot include the text “anal” in the group name.
This is particularly amusing considering Google’s close ties with a certain consumer genetics company in the bay area. We ended up going with “DNA Journal Group” instead. Thanks for keeping it clean, Google!
Tags: DNA, Fail, Funny, Google, Google Groups
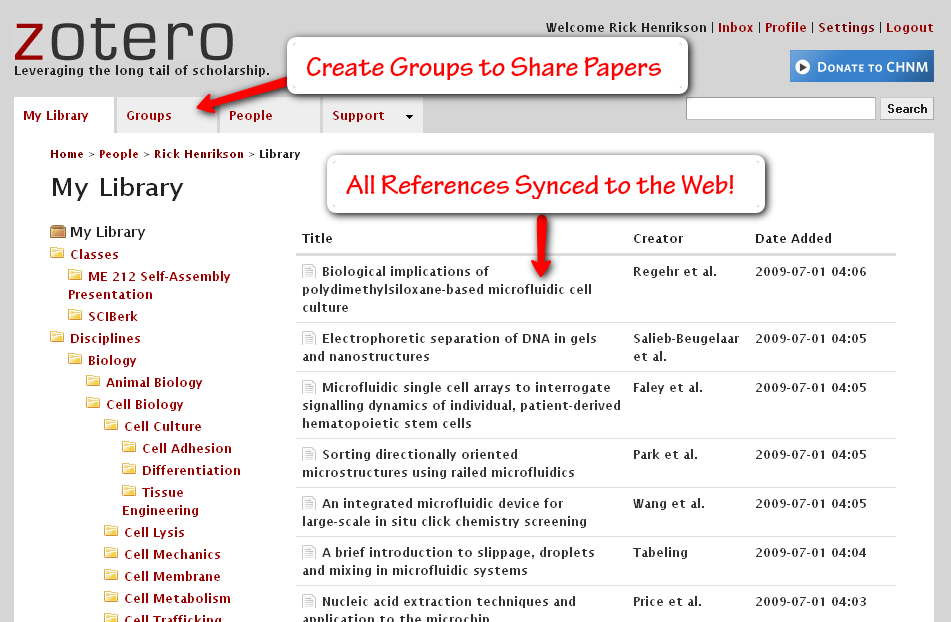
Zotero is magic for saving, organizing, and sharing documents on the web

The Citation Management Problem
Most of you probably recall the disproportionately large emphasis elementary and secondary education often placed on “the bibliography.” A significant amount of time was spent teaching you just how to cite someone else’s work in the right way. Where does the author’s name go? The edition? The page numbers? …Who cares? Now you shouldn’t have to spend any tedious hours organizing reference information thanks to awesome software that is freely available from Zotero. But it’s not just for book reports; I would highly recommend Zotero if any of the following applies to you:
- You use EndNote (and thus appreciate how much it sucks, on top of how expensive it is).
- You are involved in any stage of an academic career (student, professor, creepy old guy at the library).
- You are a professional who needs to keep abreast of current published developments in your field.
- You currently use a tool like EverNote to clip information from the web, but sometimes you want to pull out the semantic data hidden in the html (author, publication date, journal, etc.). Or maybe now you’re realizing you should.
If you’re still reading, and you haven’t already had me physically force Zotero installation upon you, then you’re about to pick up a pretty useful tool to accelerate your research. I’ve been using it since February 2008, and I’ve collected 1,778 references to date.
Zotero is Feature-Rich
Created by the Center for History and New Media at George Mason University, Zotero is an open source project that has grown rapidly in recent months. My friend over at wired liked it so much he featured it in a blog post a little over a year ago. Since then, the major release of Zotero 2.0 has allowed users to sync and share all of the reference information they have collected in online groups and folders. But let’s go through some of the key components first.
The Library
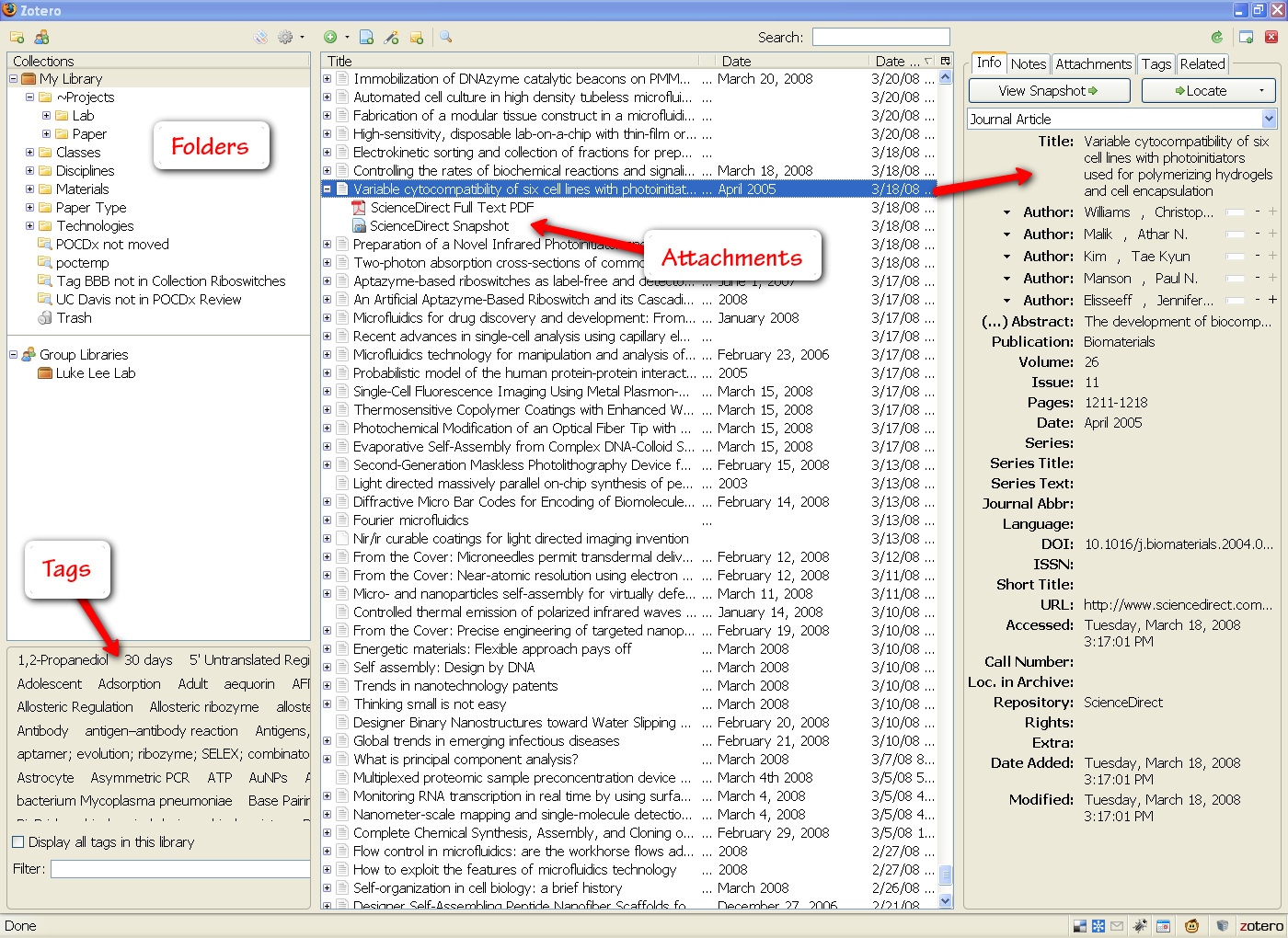
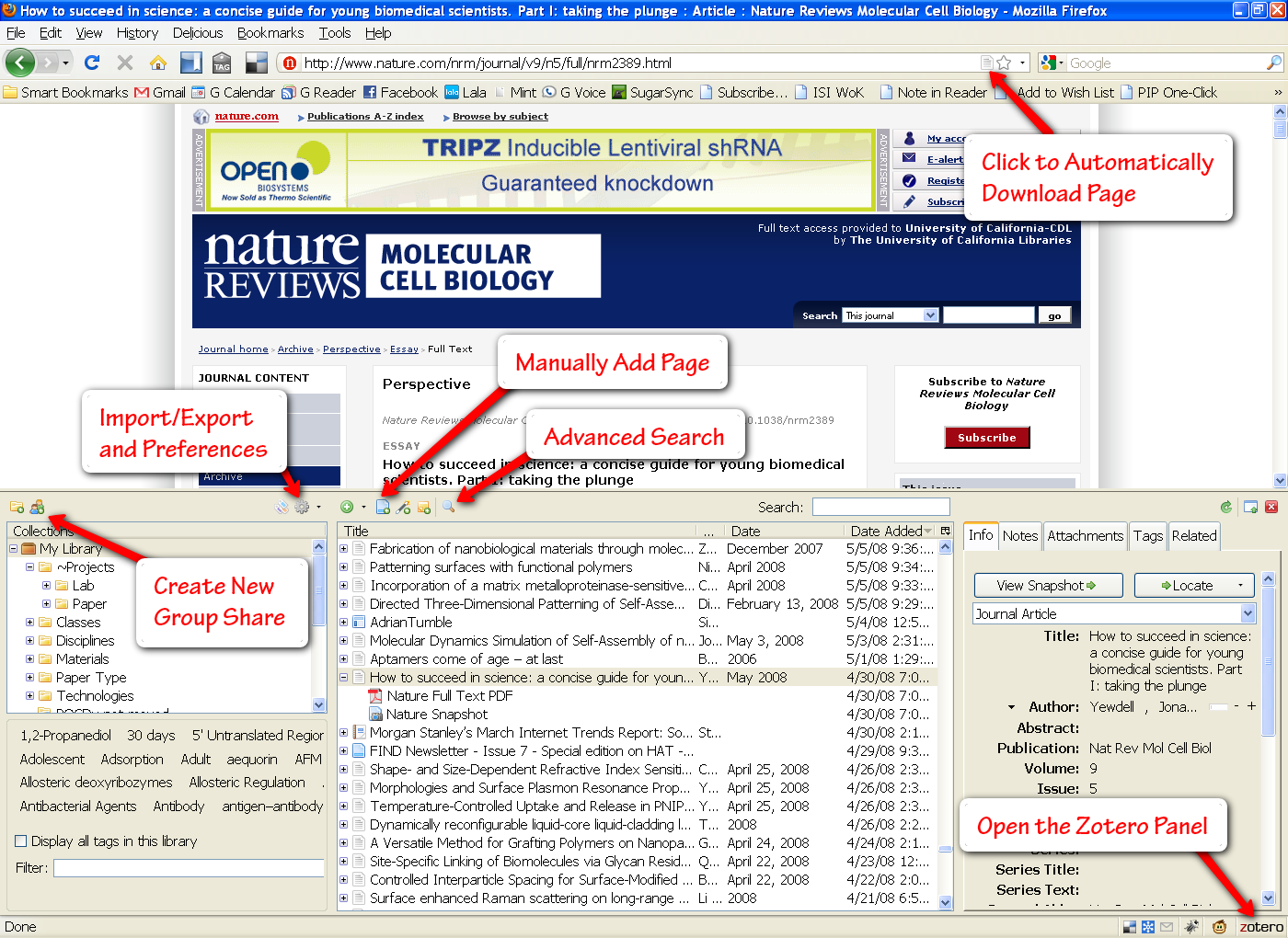
My Zotero library window expanded in Firefox (click to enlarge)
The center panel shows all of my Zotero files. Each entry can be expanded to show its associated attachments. Zotero will automatically download the html page for a file, as well as any pdf’s that are available. You can add as many more attachments and notes as you would like. This center panel can also be filtered by folder and tags (options in the left panel).
Zotero automatically reads all of the metadata associated with an entry and makes this visible in the right panel. And if for some reason Zotero is unable to pull in your document information, you can manually enter details in any field (though you can see how tedious this would be). Beside the “Info” tab, you will commonly use the “Notes” and “Tags” tabs to jot down some more details.
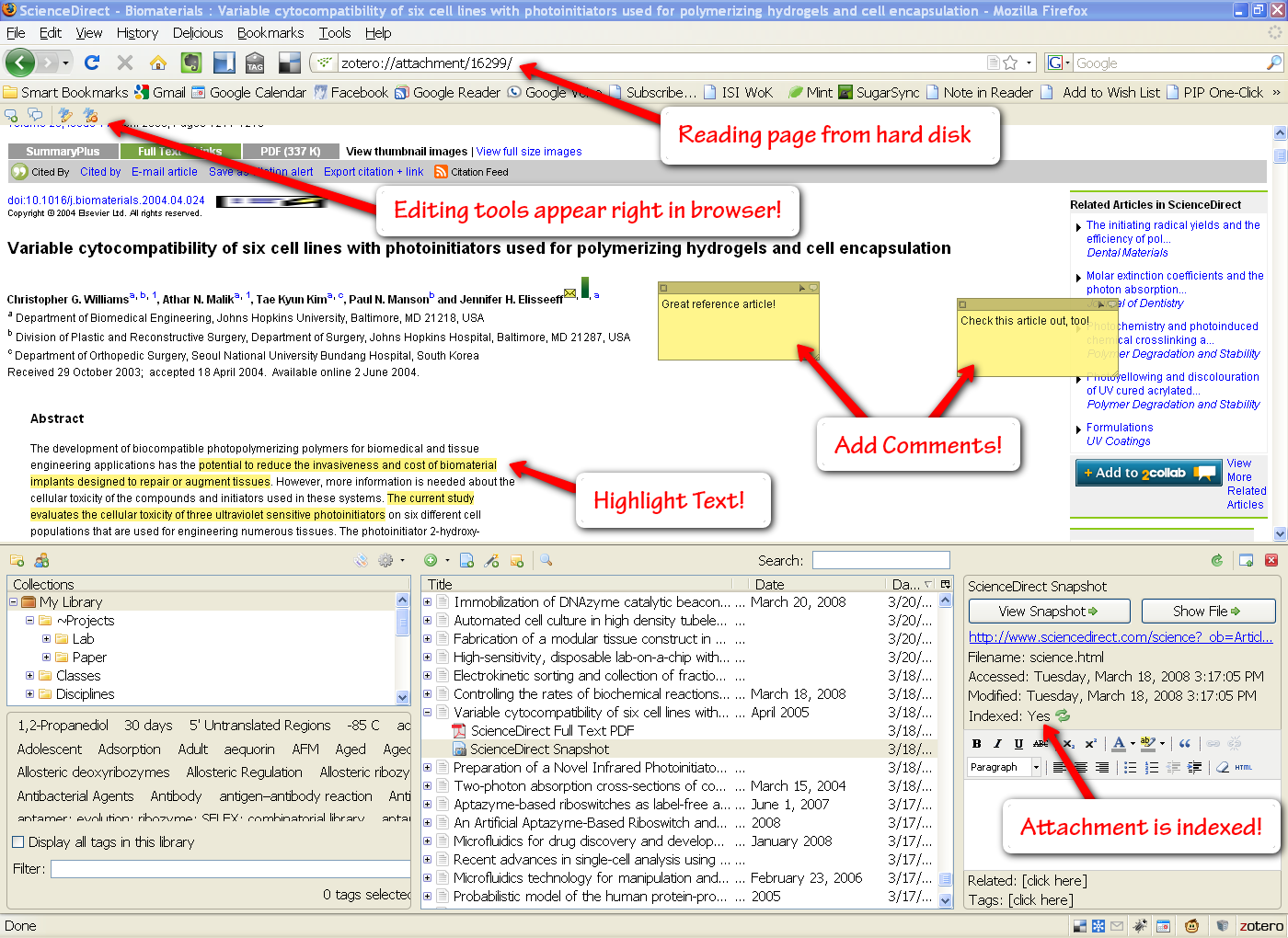
The Attachment Viewer
Zotero even lets you open attachments (html and pdf) right in the browser window. Doing so will cause a little annotation toolbar to slide out of the top of the window, allowing you to highlight text and add comment bubbles with your own notes. This is pretty useful, though I generally opt to take notes in Foxit Reader, a quick pdf reader (will add a post on that later).
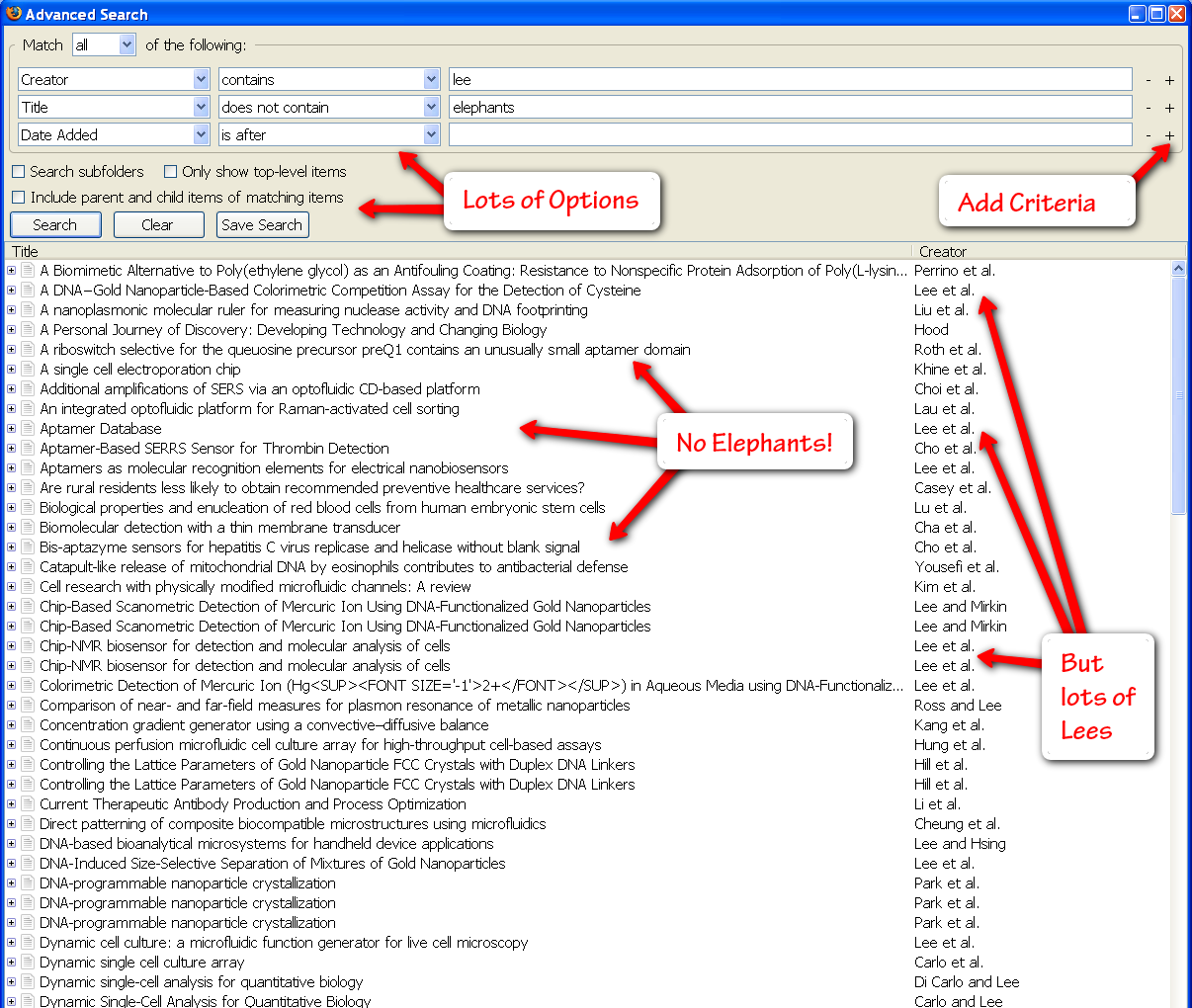
Advanced Search
One of the best things about Zotero is the rapid search. You can type any search term into the general search box above the center pane in Zotero’s main window (this will search through all of your items, including the text of all attachments such as pdf and html files). You can also click the little magnifying glass to open an advanced search with unlimited searching criteria (author, publisher, date, etc.). Having a great search interface guarantees you’ll be able to find any paper you’re looking for in your gigantic library.
The Web Interface
This is clearly the least developed area of Zotero right now, but it is nice to see that Zotero is moving towards the cloud. You can see your entire Zotero library here, which can be automatically (or manually) synced from the Firefox plugin. You can create groups for sharing files (right now it seems the only way to add files is through the plugin, though – so you’ll have to drag them into the group folder and sync them to the web to make them appear). Currently Zotero will not sync your pdf/html attachments, but they will help you do this through your own server space (it seems they will be adding a simpler, hosted, option in the future). For now I just back up my pdf files on the web using SugarSync (more on that later).
Easy to Set Up and Use
Since Zotero is a plugin for Firefox, all you have to do is make sure you have the latest version of the browser and then install the plugin directly from Zotero’s main page (I suggest using the latest version 2.0 beta – it works well and has a lot more features). After a quick Firefox restart, you should see the Zotero icon in the bottom right pane of the Firefox window. Clicking this button opens the Zotero library where you can view and edit all of your saved pages. I’d recommend adjusting a few of the preferences after a fresh install (click the gear icon at the top of the Zotero panel and select “Preferences…”):
- General Tab: I checked all of the boxes here. You’re going to want Zotero to automatically download associated pdf and html files.
- Sync Tab: Put your zotero.org login information here.
- Search Tab: Set up pdf indexing. There should be a button you can press for Zotero to automatically install a couple of pdf reader programs to enable this functionality. Now all your pdf files will be searchable.
- Advanced Tab: You can change the folder where Zotero will store all of your files. If you don’t care, you can just leave it stored in your Firefox profile directory.
Organizing Your Information
Everyone has their own way of organizing information, and fortunately Zotero covers the main options that please most people. I’m going to tell you the right way to organize things, though. Zotero gives you two main tools to organize citations: folders and tags. Folders are basically the same thing as tags (an item can exist in more than one folder), except that they (1) have a hierarchy and (2) will not be preserved if the folder is exported. You can think of folders as being like tags that are associated with your particular library, but not with the individual items (so the information gets lost when individual items are exported). Tags, on the other hand, are tied to individual citations and are thus preserved on export.
When I first started using Zotero, I thought I’d get super organized and I created a pretty thorough folder-based ontology. I could readily pull up citations in the folder Materials->Biomolecules->DNA->Aptamers. However, this required a lot of manual organization and this quickly became prohibitively draining on my time. So here is my simplified strategy for organizing citations:
- Assign to “Project” Folders. I create a folder called “Projects” with subfolders for each project I am currently working on. I often think about and use papers in terms of the project they are associated with, so this is the easiest and most convenient way to categorize papers. You can also drag multiple citations into a folder at once (which is convenient since I tend to get citations in batches that are all associated with the same projects). You could similarly have a “Classes” folder with subfolders for each class you are collecting references, and subfolders for specific assignments in any given class.
- Tag the Priority. Initially I had two main tags for this: “ToRead” and “Read”. I later realized I would have to prioritize what I read in some way, so I created a “ToReadNext” tag that I use to help guide my immediate reading, reserving the “ToRead” files for more leisurely browsing. Once I have finished reading something, I mark it as “Read” and “Annotated” (if I have made highlights and other annotations in the digital version). This makes it easy for me to quickly filter through only the papers that I’ve already finished.
- Assign to “Paper Type” Folders. There are several broad classes of paper types that are relevant to me as a researcher, so it is convenient to make a quick categorization when I pull a paper. The main categories I use are “Reviews” (for broad review articles), “Model Papers” (examples of great science and communication), and “Guidelines” (any references that describe best practice guidelines for my field), and “RFA” (any requests for applications relevant to my work – ultimately everything is all about the benjamins).
This may seem like a lot of work, but it’s actually rather quick and can save you tons of time when you are preparing a report or just updating a coworker on the most important literature in your field (it only takes me a second to consistently transfer all of the important literature to my undergraduate students). However, if you are on the lazy end of the spectrum, you can also just rely on the automatic tags generated by Zotero based on metadata on the page (in the case of scientific articles, these tags often come from the “keywords” section). Plus the built-in search is incredibly effective in finding what you’re looking for.
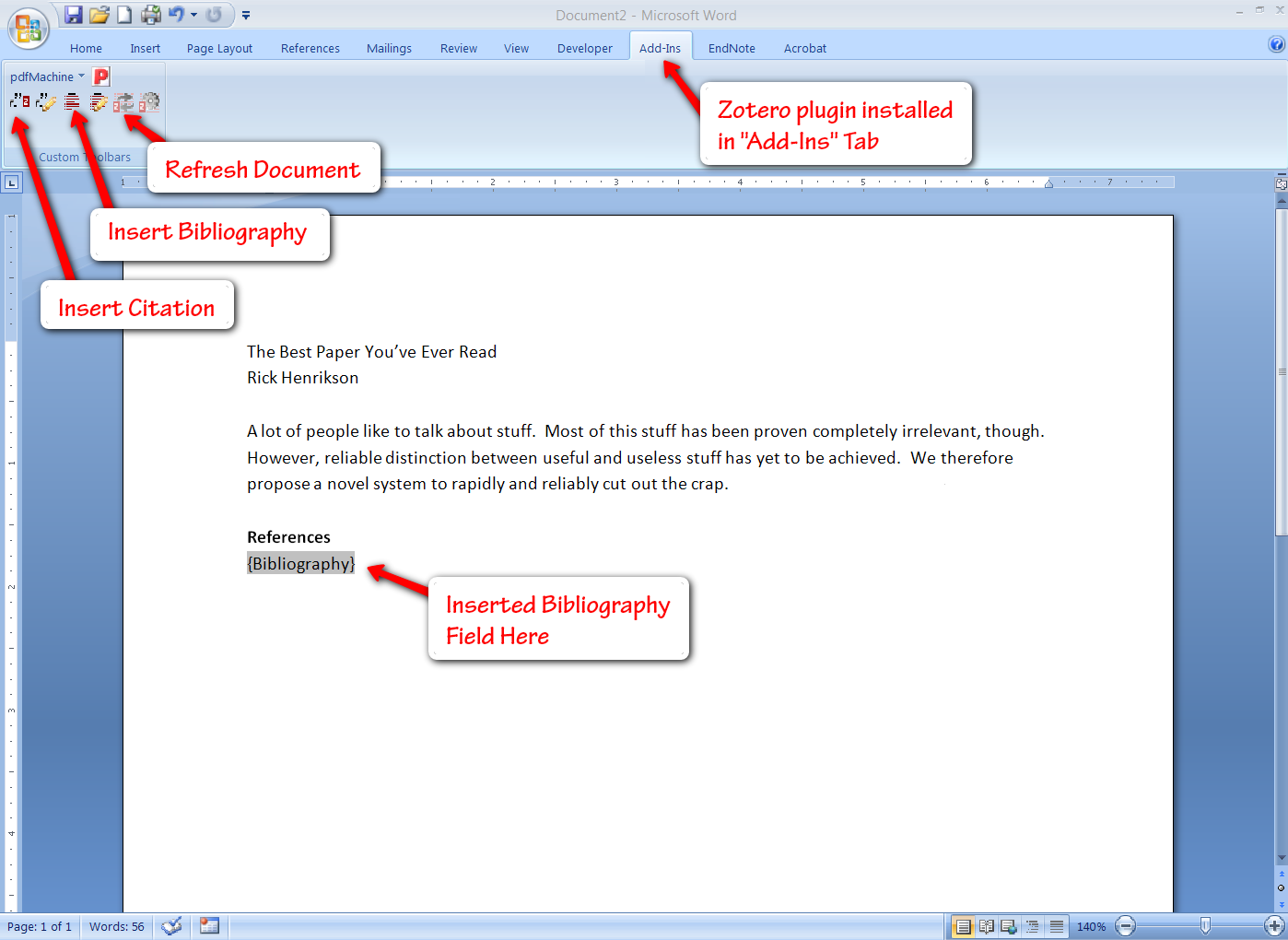
Integrated Bibliography Publishing
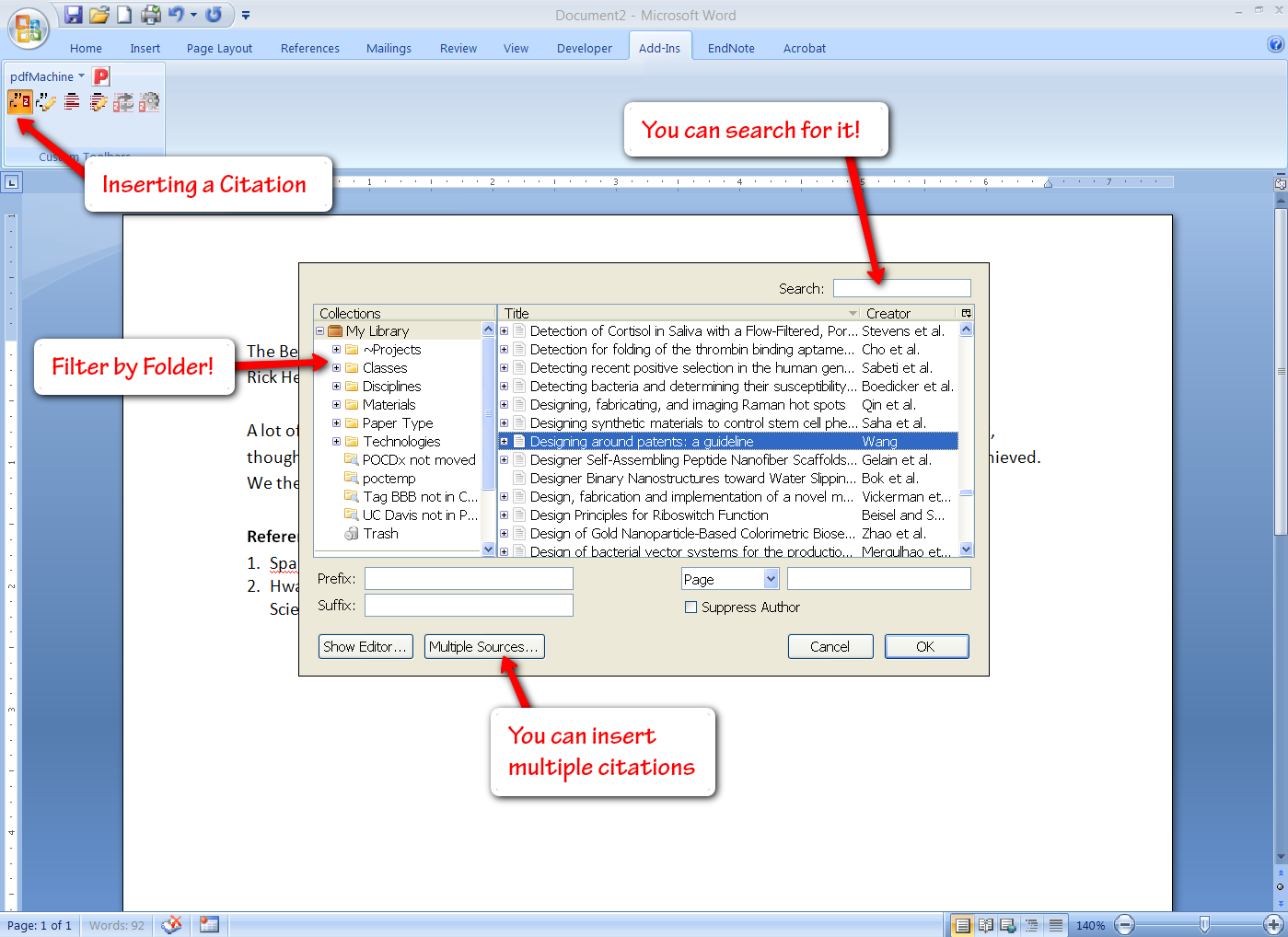
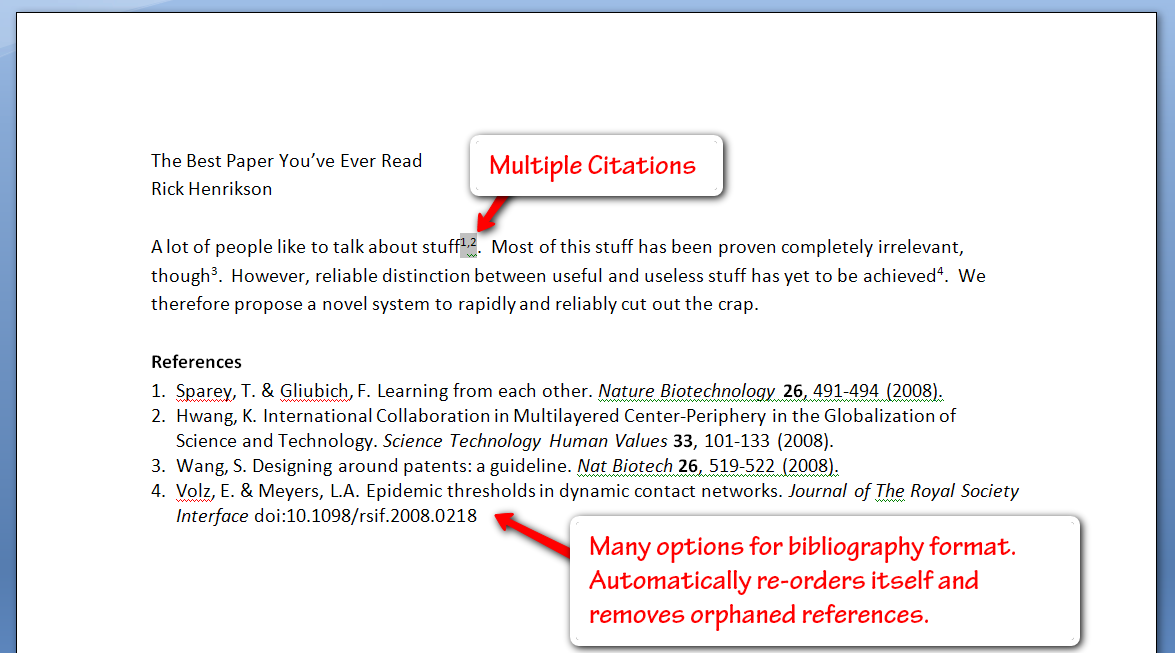
Zotero also offers plugins for Word, OpenOffice, and NeoOffice so you can readily insert citations into your documents (these options should cover all Windows & Mac users). You get a few extra buttons in the “Add-Ins” tab of Word, allowing you to insert a bibliography (you can select from a number of bibliography formats, or create your own), and subsequently insert and edit citations. Inserting citations is fairly simple, especially if you have organized them by project, and Zotero even gives you search functionality here.
The only issue here is that everything is done with fields, which can lead to some file issues at times. However, all of the citations and bibliographies you insert can be manually formatted and adjusted as you see fit (simply hitting the Zotero refresh button will bring the bibliography back to its original formatting, and also adjust citation numbering to match any changes that have been made by deleting or inserting references). This simple bibliography creator has saved me more than once in a time crunch.
Troubleshooting
Although Zotero seems to recognize most every site I want to pull citations from, I do occasionally find an obscure site that doesn’t have a Zotero translator yet. When this happens, I try to search another paper aggregation site that Zotero does recognize (such as PubMed or Google Scholar). If you still can’t find the paper on a translated site, you can either write a translator for Zotero, or you can just enter the information manually.
Sometimes Zotero will accurately scrape the metadata from a site, but it will be unable to automatically download the pdf associated with it. In these cases, you can actually drag a link to the pdf file right onto the citation in your Zotoro library and it will be linked and indexed. Alternatively, you can download the pdf manually and then use the “Attachments” tab of the library to associate an attachment with it (pdf or otherwise). Make sure to use the “Store Copy of File…” option to actually store and index the document in your Zotero database (rather than just linking out to the document wherever it exists on your hard drive).
If you run into any other issues while using Zotero, I highly recommend consulting the Zotero forums. The users and developers are generally quick to respond to any issues you post (though you should first conduct a search to make sure your questions hasn’t already been answered elsewhere).
Competitors
There are several other options for citation management. The old standard is EndNote, though the functionality and user experience clearly pale in comparison to Zotero’s free and open source alternative. Backed into a corner by an obviously superior product, EndNote even lashed out with a lawsuit that was ultimately dismissed. So unless you want to spend a lot of money on a broken product, I recommend steering clear of EndNote.
I’ve been told there is some Apple software called Papers that attempts to do similar things, albeit in an iTunes-style application. However, I generally wouldn’t recommend the use of Apple products. Also, this one seems to cost money, and it’s not clear that it can automatically pull metadata out of references you find on the web (at least from browsing their features page).
The biggest competitor to Zotero is Mendeley, a product that describes itself as a “Last.fm for references”. I tested Mendeley out when it was first available and it was a fairly crippled experience when compared with Zotero. However, it seems they have made a lot of progress since then, particularly on the social front, so I will give them another chance and report back shortly. Their system seems to be built around pdf’s to a large extent, so if you have a huge pdf library it might be worth checking them out.
I spoke with an employee from Mendeley at the most recent SciBarCamp in Palo Alto and he said Zotero is a great product (he even uses it from time to time), but he’s excited about the social/recommendation features that Mendeley has in the works. All of their current features will remain free, but they will be introducing some premium features that will require some moolah. It’ll be interesting to see how this plays out, though I’d place my bet on the free and open source solution.
The Bottom Line
Zotero an awesome and simple tool for managing and sharing any documents you come across on the web (automatically storing bibliographic metadata as well as any associated html and pdf files). It will change your life if you regularly interact with publications online that you later need to reference. It’s even supported by Berkeley.
Tags: Citations, EndNote, Mozilla Firefox, Zotero

![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=b4860935-7265-4836-86d3-6d2f5eb003cf)



![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=cb61c8af-10cb-4b74-b0d9-9a2b7eb440a9)
![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=55721b5e-d4f6-4b4a-be43-6d07f0ac4adc)
![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=78bd225a-4306-4a17-83f0-cf214579cb9e)
![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=159033e3-0568-4f46-925a-8f4467a7854d)